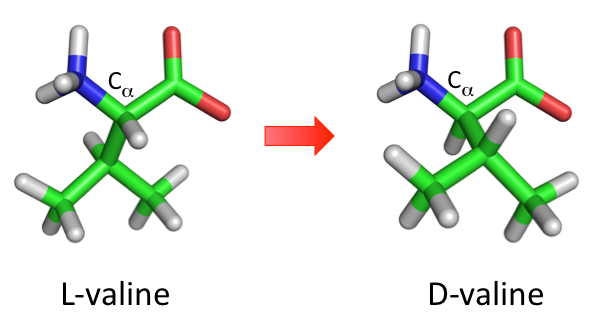
D-amino acids are mirror images of L-amino acids, where the chirality at carbon alpha has been inverted (see figure below). They rarely occur naturally in organisms except for some bacteria. D-amino acids are highly resistant to protease mediated degradation and have a low immunogenic response. This makes D-amino acids interesting candidates for peptidomimetics drug design.
Here, we provide all data to work with both natural and non-natural D-amino acids.
All D-amino acids can be accessed from this page by selecting the corresponding L-form.
Non-natural D-amino acids with an existing 3-letter PDB code for the L-form have been named by adding a D in front of the code (ex: NAL -> DNAL). Other non-natural D-amino acids with a 4-letter code for the L-form have been given a 4-letter code starting with D and reminiscent of the L-form (ex: ADAM -> DADM).
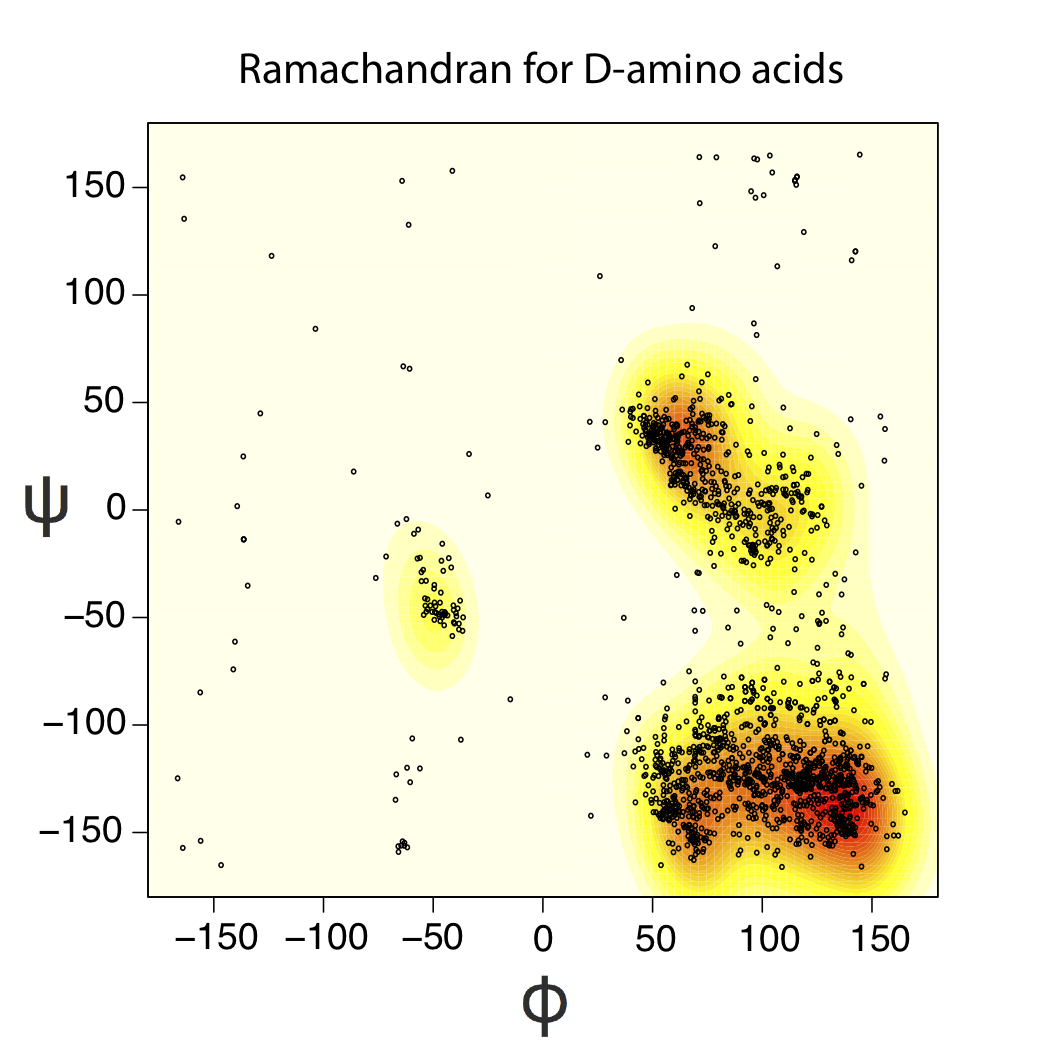
You can also insert D-amino acids into existing structures using our visualization plugins. We note that depending on the backbone dihedral angles (φ and ψ), inserting D-sidechains may result in significant changes in the backbone conformations because of steric clashes. Below we show the portion of the ramachandran diagram that is sampled by D-amino acids in experimental crystal structures.